Monday Poster Session
Category: Liver
P2409 - Metabolomic Features Improve Liver Stiffness Measurement Prediction in Patients With Nonalcoholic Fatty Liver Disease
Monday, October 23, 2023
10:30 AM - 4:15 PM PT
Location: Exhibit Hall
Has Audio
.jpg)
Brent J. Gawey, MD, MSc
Mayo Clinic
Rochester, MN
Presenting Author(s)
Brent J. Gawey, MD, MSc1, Andrew ElHabr, PhD2, Matthew R. Smith, PhD3, Thomas R. Ziegler, MD, MSc3, Dean P. Jones, PhD3, Andrew S. Neish, MD3
1Mayo Clinic, Rochester, MN; 2Value Analytics Labs, Austin, TX; 3Emory University School of Medicine, Atlanta, GA
Introduction: Nonalcoholic fatty liver disease (NAFLD) is widely prevalent with cases expected to increase in coming years. At present, liver stiffness measurement (LSM) is a commonly used test, typically performed using transient elastography, to assess the degree of fibrosis in the liver. With the expected rise in NAFLD, the need for more cost-effective and widely available diagnostic tests are needed. In recent years, high resolution metabolomics (HRM), a platform allowing for profiling of metabolic features in biosamples, has seen significant advancements. HRM is relatively low cost and can be widely implemented across clinical environments. In this study, we assessed model performance for predicting LSM using base demographic models and those enhanced with metabolomic features.
Methods: Fasting plasma from 139 participants with pre-existing liver fibrosis (mean age 51.1 ± 12.4 years, 66% female) was analyzed using HRM. Liver fibrosis was quantified using liver stiffness measurement (LSM) scores. A base linear regression model to predict LSM was constructed using age, sex, body mass index (BMI), and presence of diabetes. Metabolomic features were then iteratively integrated into the base model and changes in in-sample goodness of fit of the models was assessed using likelihood ratio tests using p-values adjusted for multiple comparisons. In addition, 100 bootstrap samples of the data were used to fit linear regression models to predict LSM using the same set of base variables. Subsequently, these models were augmented by incorporating metabolomic features. The predictive performance of the base models versus the models enhanced with metabolic features was compared using out-of-sample predictions.
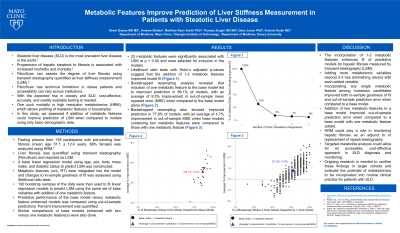
Results: Likelihood ratio tests suggest that the addition of 1-2 metabolomic features could improve model fit. Bootstrapped resampling analysis then revealed that including a metabolomic feature led to improved prediction in 86% of samples with an average 9% reduction in out-of-sample mean squared error.
Discussion: This study shows that incorporation of metabolomic features enhances predictive models for hepatic fibrosis using LSM. Incorporating any single metabolomic feature among numerous candidates improved both in-sample goodness of fit and out-of-sample prediction error when compared to a base model. Ongoing research is needed to confirm these findings and evaluate their potential to change management in NAFLD.
Disclosures:
Brent J. Gawey, MD, MSc1, Andrew ElHabr, PhD2, Matthew R. Smith, PhD3, Thomas R. Ziegler, MD, MSc3, Dean P. Jones, PhD3, Andrew S. Neish, MD3. P2409 - Metabolomic Features Improve Liver Stiffness Measurement Prediction in Patients With Nonalcoholic Fatty Liver Disease, ACG 2023 Annual Scientific Meeting Abstracts. Vancouver, BC, Canada: American College of Gastroenterology.
1Mayo Clinic, Rochester, MN; 2Value Analytics Labs, Austin, TX; 3Emory University School of Medicine, Atlanta, GA
Introduction: Nonalcoholic fatty liver disease (NAFLD) is widely prevalent with cases expected to increase in coming years. At present, liver stiffness measurement (LSM) is a commonly used test, typically performed using transient elastography, to assess the degree of fibrosis in the liver. With the expected rise in NAFLD, the need for more cost-effective and widely available diagnostic tests are needed. In recent years, high resolution metabolomics (HRM), a platform allowing for profiling of metabolic features in biosamples, has seen significant advancements. HRM is relatively low cost and can be widely implemented across clinical environments. In this study, we assessed model performance for predicting LSM using base demographic models and those enhanced with metabolomic features.
Methods: Fasting plasma from 139 participants with pre-existing liver fibrosis (mean age 51.1 ± 12.4 years, 66% female) was analyzed using HRM. Liver fibrosis was quantified using liver stiffness measurement (LSM) scores. A base linear regression model to predict LSM was constructed using age, sex, body mass index (BMI), and presence of diabetes. Metabolomic features were then iteratively integrated into the base model and changes in in-sample goodness of fit of the models was assessed using likelihood ratio tests using p-values adjusted for multiple comparisons. In addition, 100 bootstrap samples of the data were used to fit linear regression models to predict LSM using the same set of base variables. Subsequently, these models were augmented by incorporating metabolomic features. The predictive performance of the base models versus the models enhanced with metabolic features was compared using out-of-sample predictions.
Results: Likelihood ratio tests suggest that the addition of 1-2 metabolomic features could improve model fit. Bootstrapped resampling analysis then revealed that including a metabolomic feature led to improved prediction in 86% of samples with an average 9% reduction in out-of-sample mean squared error.
Discussion: This study shows that incorporation of metabolomic features enhances predictive models for hepatic fibrosis using LSM. Incorporating any single metabolomic feature among numerous candidates improved both in-sample goodness of fit and out-of-sample prediction error when compared to a base model. Ongoing research is needed to confirm these findings and evaluate their potential to change management in NAFLD.
Disclosures:
Brent Gawey indicated no relevant financial relationships.
Andrew ElHabr indicated no relevant financial relationships.
Matthew Smith indicated no relevant financial relationships.
Thomas Ziegler indicated no relevant financial relationships.
Dean Jones indicated no relevant financial relationships.
Andrew Neish indicated no relevant financial relationships.
Brent J. Gawey, MD, MSc1, Andrew ElHabr, PhD2, Matthew R. Smith, PhD3, Thomas R. Ziegler, MD, MSc3, Dean P. Jones, PhD3, Andrew S. Neish, MD3. P2409 - Metabolomic Features Improve Liver Stiffness Measurement Prediction in Patients With Nonalcoholic Fatty Liver Disease, ACG 2023 Annual Scientific Meeting Abstracts. Vancouver, BC, Canada: American College of Gastroenterology.

