Monday Poster Session
Category: Stomach
P2763 - Unveiling the Power of Molecular Testing in Detecting Clarithromycin-Resistant Helicobacter pylori in Stool: A Systematic Review and Meta-Analysis
Monday, October 23, 2023
10:30 AM - 4:15 PM PT
Location: Exhibit Hall

Ahmed Salih, MD
Indiana University School of Medicine
Muncie, IN
Presenting Author(s)
Award: Presidential Poster Award
Ahmed Salih, MD1, Marriam Ali, MD1, Sasmith Menakuru, MD1, Vijaypal Dhillon, MD1, Ayman Elawad, MD2, Fouad Jaber, MD3, Fernando Stancampiano, MD4, Dana Harris, MD4, Yan Bi, MD, PhD4
1Indiana University School of Medicine, Muncie, IN; 2Howard University Hospital, Washington, DC; 3University of Missouri-Kansas City, Kansas City, MO; 4Mayo Clinic, Jacksonville, FL
Introduction: The increasing prevalence of clarithromycin (CLA)-resistant H. pylori strains poses a significant challenge in the management of H. pylori infections. This meta-analysis investigates the diagnostic accuracy of molecular testing methods for identifying CLA-resistant H. pylori strains in stool.
Methods: A systematic search was conducted in PubMed, Ovid, EMBASE, and Cochrane databases up to April 2023. It aimed to identify full-text studies evaluating the efficacy of molecular stool tests in detecting CLA-resistant H. pylori. Inclusion criteria specified the adult population ( >18 years old), molecular stool testing for CLA resistance, and comparison with antibiotic susceptibility or PCR on gastric biopsy. Studies involving pediatric populations, alternative methods to PCR or stool samples, and reference tests other than gastric biopsy were excluded. The bivariate model was employed to analyze the diagnostic accuracy data and estimate summary measures.
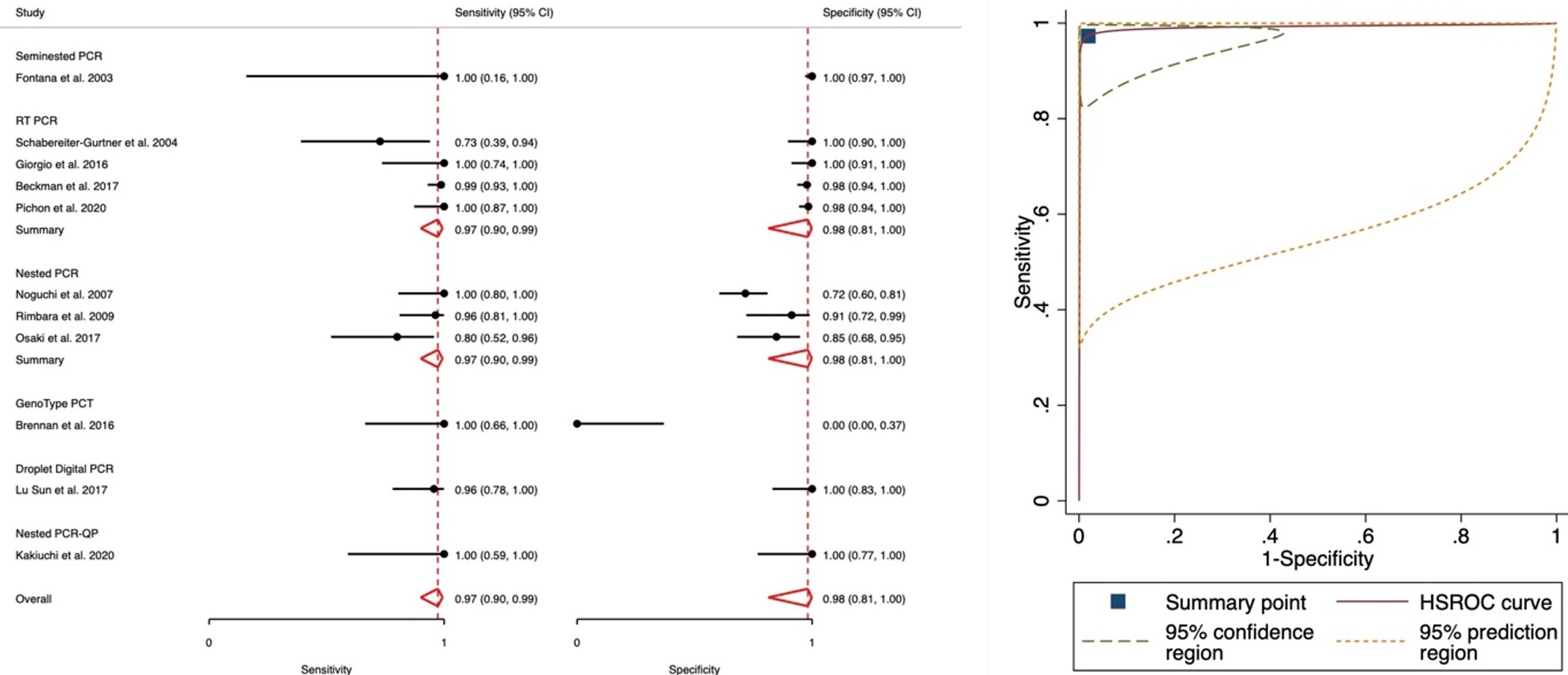
Results: The initial search yielded a total of 1832 studies, from which 121 relevant articles were selected based on title and abstract review. Eleven studies met the inclusion criteria. A total of 866 patients were included in the analysis. In all studies, except two, a culture of the gastric biopsy served as the gold standard, while PCR on the biopsy was used in the remaining two. Table 1 provides detailed characteristics of each study.
The analysis of sensitivity and specificity values revealed a sensitivity range of 0.73 to 1.0, with a pooled estimate of 0.97 (95% CI: 0.9-0.99, I2=15.52). The specificity ranged from 0 to 1.0, with a pooled estimate of 0.98 (95% CI: 0.81-1.0, I2=1.48). The observed heterogeneity between studies was minimal. The forest plot and the summary of receiver operator curve are detailed in Figure 1.
Discussion: Our meta-analysis provides strong evidence for the effectiveness of molecular testing methods in accurately detecting clarithromycin-resistant H. pylori strains in stool samples. The high sensitivity and specificity observed across the studies support their reliability in clinical practice, particularly in outpatient settings, where invasive procedures can be minimized. Additionally, the implementation of these methods can have notable cost-effectiveness implications. In summary, our findings highlight the promise of molecular testing for identifying clarithromycin resistance in H. pylori, offering opportunities for tailored treatment strategies and improved patient outcomes.
Disclosures:
Ahmed Salih, MD1, Marriam Ali, MD1, Sasmith Menakuru, MD1, Vijaypal Dhillon, MD1, Ayman Elawad, MD2, Fouad Jaber, MD3, Fernando Stancampiano, MD4, Dana Harris, MD4, Yan Bi, MD, PhD4. P2763 - Unveiling the Power of Molecular Testing in Detecting Clarithromycin-Resistant Helicobacter pylori in Stool: A Systematic Review and Meta-Analysis, ACG 2023 Annual Scientific Meeting Abstracts. Vancouver, BC, Canada: American College of Gastroenterology.
Ahmed Salih, MD1, Marriam Ali, MD1, Sasmith Menakuru, MD1, Vijaypal Dhillon, MD1, Ayman Elawad, MD2, Fouad Jaber, MD3, Fernando Stancampiano, MD4, Dana Harris, MD4, Yan Bi, MD, PhD4
1Indiana University School of Medicine, Muncie, IN; 2Howard University Hospital, Washington, DC; 3University of Missouri-Kansas City, Kansas City, MO; 4Mayo Clinic, Jacksonville, FL
Introduction: The increasing prevalence of clarithromycin (CLA)-resistant H. pylori strains poses a significant challenge in the management of H. pylori infections. This meta-analysis investigates the diagnostic accuracy of molecular testing methods for identifying CLA-resistant H. pylori strains in stool.
Methods: A systematic search was conducted in PubMed, Ovid, EMBASE, and Cochrane databases up to April 2023. It aimed to identify full-text studies evaluating the efficacy of molecular stool tests in detecting CLA-resistant H. pylori. Inclusion criteria specified the adult population ( >18 years old), molecular stool testing for CLA resistance, and comparison with antibiotic susceptibility or PCR on gastric biopsy. Studies involving pediatric populations, alternative methods to PCR or stool samples, and reference tests other than gastric biopsy were excluded. The bivariate model was employed to analyze the diagnostic accuracy data and estimate summary measures.
Results: The initial search yielded a total of 1832 studies, from which 121 relevant articles were selected based on title and abstract review. Eleven studies met the inclusion criteria. A total of 866 patients were included in the analysis. In all studies, except two, a culture of the gastric biopsy served as the gold standard, while PCR on the biopsy was used in the remaining two. Table 1 provides detailed characteristics of each study.
The analysis of sensitivity and specificity values revealed a sensitivity range of 0.73 to 1.0, with a pooled estimate of 0.97 (95% CI: 0.9-0.99, I2=15.52). The specificity ranged from 0 to 1.0, with a pooled estimate of 0.98 (95% CI: 0.81-1.0, I2=1.48). The observed heterogeneity between studies was minimal. The forest plot and the summary of receiver operator curve are detailed in Figure 1.
Discussion: Our meta-analysis provides strong evidence for the effectiveness of molecular testing methods in accurately detecting clarithromycin-resistant H. pylori strains in stool samples. The high sensitivity and specificity observed across the studies support their reliability in clinical practice, particularly in outpatient settings, where invasive procedures can be minimized. Additionally, the implementation of these methods can have notable cost-effectiveness implications. In summary, our findings highlight the promise of molecular testing for identifying clarithromycin resistance in H. pylori, offering opportunities for tailored treatment strategies and improved patient outcomes.

Figure: Figure 1
Disclosures:
Ahmed Salih indicated no relevant financial relationships.
Marriam Ali indicated no relevant financial relationships.
Sasmith Menakuru indicated no relevant financial relationships.
Vijaypal Dhillon indicated no relevant financial relationships.
Ayman Elawad indicated no relevant financial relationships.
Fouad Jaber indicated no relevant financial relationships.
Fernando Stancampiano indicated no relevant financial relationships.
Dana Harris indicated no relevant financial relationships.
Yan Bi indicated no relevant financial relationships.
Ahmed Salih, MD1, Marriam Ali, MD1, Sasmith Menakuru, MD1, Vijaypal Dhillon, MD1, Ayman Elawad, MD2, Fouad Jaber, MD3, Fernando Stancampiano, MD4, Dana Harris, MD4, Yan Bi, MD, PhD4. P2763 - Unveiling the Power of Molecular Testing in Detecting Clarithromycin-Resistant Helicobacter pylori in Stool: A Systematic Review and Meta-Analysis, ACG 2023 Annual Scientific Meeting Abstracts. Vancouver, BC, Canada: American College of Gastroenterology.


