Sunday Poster Session
Category: Colorectal Cancer Prevention
P0359 - Targeting the Sugar Code: Serum Glycoproteome Profiling for Early Detection of Advanced Adenoma and Colorectal Cancer
Sunday, October 22, 2023
3:30 PM - 7:00 PM PT
Location: Exhibit Hall
Has Audio
- DC
Dharini Chandrasekar, DO, MPH
InterVenn Biosciences
South San Francisco, CA
Presenting Author(s)
Award: Presidential Poster Award
Julia J.. Liu, MD1, Samir Gupta, MD, MSCS2, Aasma Shaukat, MD3, Nik Sheng Ding, MBBS, PhD4, Heiko Pohl, MD5, Khushbu Desai, MS6, Dharini Chandrasekar, DO, MPH6, Christina Guerrier, MBA6, Prasanna Ramachandran, PhD6, Karina Islas-Rios, BSc, PhD7, Richard D. Shipman, BSc8, Tomislav Čaval, BSc, MSc, PhD6, Matthew P. Campbell, PhD9, Chirag Dhar, MD, MAS10, Gege Xu, PhD6, Flavio Schwarz, PhD6, Daniel Serie, MS6, Evelien Dekker, MD, PhD11, Michael B. Wallace, MD, MPH12, Daniel Hommes, MD, PhD6, Folasade P.. May, MD, PhD, MPhil13
1Morehouse School of Medicine, Atlanta, GA; 2Jennifer Moreno Veteran Affairs San Diego Healthcare System, San Diego, CA; 3NYU Langone Health, New York, NY; 4St. Vincent’s Hospital, East Melbourne, Victoria, Australia; 5Dartmouth, Lebanon, NH; 6InterVenn Biosciences, South San Francisco, CA; 7Intervenn Biosciences, Carnegie, Victoria, Australia; 8InterVenn BioSciences, South San Francisco, CA; 9InterVenn Biosciences, Melbourne, Victoria, Australia; 10InterVenn Biosciences, San Mateo, CA; 11Amsterdam University Medical Center, Amsterdam, Noord-Holland, Netherlands; 12Sheikh Shakbout Medical City, Abudhabi, United Arab Emirates/Mayo Clinic, Jacksonville, FL, Abu Dhabi, Abu Dhabi, United Arab Emirates; 13UCLA Health, Los Angeles, CA
Introduction: Colorectal cancer (CRC) is a leading cause of cancer-related death. Immune responses within the colonic crypt have been found to contribute to CRC development. Abnormal protein glycosylation has been linked to the malignant transformation process. This study leverages stable circulating glycoproteomic markers as a means of identifying advanced adenomas (AAs) and CRC at an early stage.
Methods: Prospective (NCT05445570) and biorepository samples (to supplement AA/CRC cases) from multiple sites were included. AA was defined as polyps ≥1 cm, any size polyp with >25% villous features or high-grade dysplasia. Utilizing a glycoproteomic profiling platform that combines liquid-chromatography/mass-spectrometry and artificial-intelligence-powered data processing, we assessed glycopeptide (GP) and non-glycosylated peptide quantification transitions in peripheral blood serum. The samples were split into training, validation and hold-out testing sets. Statistical analyses were performed on normalized data from the optimized assay to develop and validate a classifier to predict probability of AA/CRCs against controls. Primary outcomes for the validation test-set were sensitivity for AA and CRC, as well as AUC for AA/CRC combined.
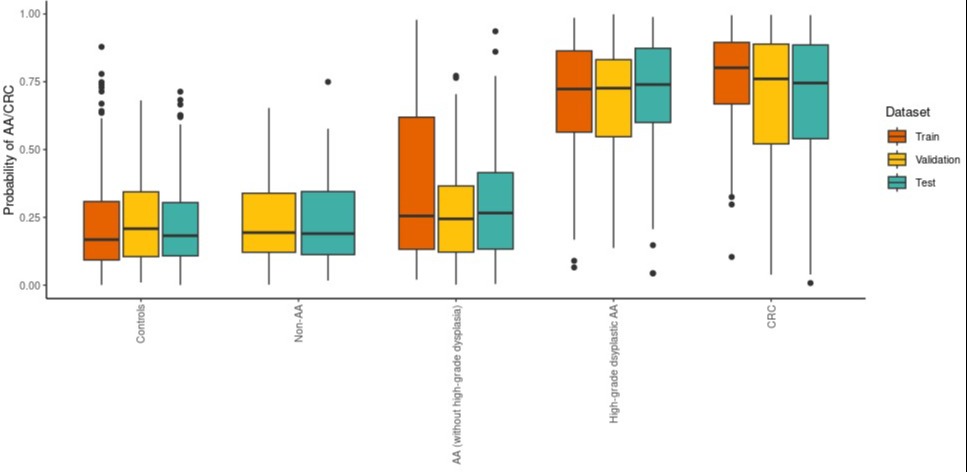
Results: We analyzed 1,356 prospectively collected samples and 681 biorepository samples: Group 1: 545 CRC (27%); Group 2: 383 AAs (19%); Group 3: 154 non-AAs (8%); Group 4: 955 colonoscopy negative controls (47%). We identified 84, 89, and 16 GPs/peptides with statistically significant abundance differences (FDR < 0.001), when comparing CRCs with controls, high-grade dysplastic (HGD) AAs with controls, and AAs to non-AAs respectively. A subset of 24 of these biomarkers were used to generate a Multivariable Classifier Model using the training and validation data sets. When the Classifier was applied to the test set it yielded an area under the receiver-operating characteristic of 0.83 for the detection of AA and CRC. Using a defined cutoff, the Classifier sensitivity for all CRC stages was 80.9% (85.0% stage 1&2); for HGDs 89.6% and for all AAs 43.8% with specificity of 90.4% for controls and 89.6% for non-AAs in the test set.
Discussion: Using glycoproteomic profiling, we detected glycosylation changes associated with the host immune response to AAs with HGD and CRC. The high sensitivity and specificity of this strategy supports the development of a non-invasive blood-based screening test that complements current screening strategies.
Disclosures:
Julia J.. Liu, MD1, Samir Gupta, MD, MSCS2, Aasma Shaukat, MD3, Nik Sheng Ding, MBBS, PhD4, Heiko Pohl, MD5, Khushbu Desai, MS6, Dharini Chandrasekar, DO, MPH6, Christina Guerrier, MBA6, Prasanna Ramachandran, PhD6, Karina Islas-Rios, BSc, PhD7, Richard D. Shipman, BSc8, Tomislav Čaval, BSc, MSc, PhD6, Matthew P. Campbell, PhD9, Chirag Dhar, MD, MAS10, Gege Xu, PhD6, Flavio Schwarz, PhD6, Daniel Serie, MS6, Evelien Dekker, MD, PhD11, Michael B. Wallace, MD, MPH12, Daniel Hommes, MD, PhD6, Folasade P.. May, MD, PhD, MPhil13. P0359 - Targeting the Sugar Code: Serum Glycoproteome Profiling for Early Detection of Advanced Adenoma and Colorectal Cancer, ACG 2023 Annual Scientific Meeting Abstracts. Vancouver, BC, Canada: American College of Gastroenterology.
Julia J.. Liu, MD1, Samir Gupta, MD, MSCS2, Aasma Shaukat, MD3, Nik Sheng Ding, MBBS, PhD4, Heiko Pohl, MD5, Khushbu Desai, MS6, Dharini Chandrasekar, DO, MPH6, Christina Guerrier, MBA6, Prasanna Ramachandran, PhD6, Karina Islas-Rios, BSc, PhD7, Richard D. Shipman, BSc8, Tomislav Čaval, BSc, MSc, PhD6, Matthew P. Campbell, PhD9, Chirag Dhar, MD, MAS10, Gege Xu, PhD6, Flavio Schwarz, PhD6, Daniel Serie, MS6, Evelien Dekker, MD, PhD11, Michael B. Wallace, MD, MPH12, Daniel Hommes, MD, PhD6, Folasade P.. May, MD, PhD, MPhil13
1Morehouse School of Medicine, Atlanta, GA; 2Jennifer Moreno Veteran Affairs San Diego Healthcare System, San Diego, CA; 3NYU Langone Health, New York, NY; 4St. Vincent’s Hospital, East Melbourne, Victoria, Australia; 5Dartmouth, Lebanon, NH; 6InterVenn Biosciences, South San Francisco, CA; 7Intervenn Biosciences, Carnegie, Victoria, Australia; 8InterVenn BioSciences, South San Francisco, CA; 9InterVenn Biosciences, Melbourne, Victoria, Australia; 10InterVenn Biosciences, San Mateo, CA; 11Amsterdam University Medical Center, Amsterdam, Noord-Holland, Netherlands; 12Sheikh Shakbout Medical City, Abudhabi, United Arab Emirates/Mayo Clinic, Jacksonville, FL, Abu Dhabi, Abu Dhabi, United Arab Emirates; 13UCLA Health, Los Angeles, CA
Introduction: Colorectal cancer (CRC) is a leading cause of cancer-related death. Immune responses within the colonic crypt have been found to contribute to CRC development. Abnormal protein glycosylation has been linked to the malignant transformation process. This study leverages stable circulating glycoproteomic markers as a means of identifying advanced adenomas (AAs) and CRC at an early stage.
Methods: Prospective (NCT05445570) and biorepository samples (to supplement AA/CRC cases) from multiple sites were included. AA was defined as polyps ≥1 cm, any size polyp with >25% villous features or high-grade dysplasia. Utilizing a glycoproteomic profiling platform that combines liquid-chromatography/mass-spectrometry and artificial-intelligence-powered data processing, we assessed glycopeptide (GP) and non-glycosylated peptide quantification transitions in peripheral blood serum. The samples were split into training, validation and hold-out testing sets. Statistical analyses were performed on normalized data from the optimized assay to develop and validate a classifier to predict probability of AA/CRCs against controls. Primary outcomes for the validation test-set were sensitivity for AA and CRC, as well as AUC for AA/CRC combined.
Results: We analyzed 1,356 prospectively collected samples and 681 biorepository samples: Group 1: 545 CRC (27%); Group 2: 383 AAs (19%); Group 3: 154 non-AAs (8%); Group 4: 955 colonoscopy negative controls (47%). We identified 84, 89, and 16 GPs/peptides with statistically significant abundance differences (FDR < 0.001), when comparing CRCs with controls, high-grade dysplastic (HGD) AAs with controls, and AAs to non-AAs respectively. A subset of 24 of these biomarkers were used to generate a Multivariable Classifier Model using the training and validation data sets. When the Classifier was applied to the test set it yielded an area under the receiver-operating characteristic of 0.83 for the detection of AA and CRC. Using a defined cutoff, the Classifier sensitivity for all CRC stages was 80.9% (85.0% stage 1&2); for HGDs 89.6% and for all AAs 43.8% with specificity of 90.4% for controls and 89.6% for non-AAs in the test set.
Discussion: Using glycoproteomic profiling, we detected glycosylation changes associated with the host immune response to AAs with HGD and CRC. The high sensitivity and specificity of this strategy supports the development of a non-invasive blood-based screening test that complements current screening strategies.

Figure: Figure 1. Distributions of predicted probabilities in each phenotype, stratified by dataset.
Disclosures:
Julia Liu indicated no relevant financial relationships.
Samir Gupta: cellmax llc – Consultant. epigenomics – Grant/Research Support. freenome – Grant/Research Support. geneoscopy – Consultant. guardant health – Consultant. universal diagnositics – Consultant.
Aasma Shaukat indicated no relevant financial relationships.
Nik Sheng Ding indicated no relevant financial relationships.
Heiko Pohl: InterVenn – Advisory Committee/Board Member.
Khushbu Desai: InterVenn Biosciences – Employee, Stock Options.
Dharini Chandrasekar: InterVenn Biosciences – Employee.
Christina Guerrier: InterVenn Biosciences – Employee.
Prasanna Ramachandran indicated no relevant financial relationships.
Karina Islas-Rios indicated no relevant financial relationships.
Richard Shipman: InterVenn BioSciences – Employee.
Tomislav Čaval indicated no relevant financial relationships.
Matthew Campbell indicated no relevant financial relationships.
Chirag Dhar: InterVenn Biosciences – Employee.
Gege Xu indicated no relevant financial relationships.
Flavio Schwarz indicated no relevant financial relationships.
Daniel Serie indicated no relevant financial relationships.
Evelien Dekker: Ambu – Advisor or Review Panel Member. FujiFilm – Advisor or Review Panel Member, Grant/Research Support, Speakers Bureau. GI Supply – Speakers Bureau. InterVenn – Advisory Committee/Board Member. IPSEN – Speakers Bureau. Norgine – Speakers Bureau. Olympus – Advisor or Review Panel Member, Speakers Bureau. PAION – Speakers Bureau.
Michael B. Wallace: Intervenn – Consultant.
Daniel Hommes: InterVenn Biosciences – Employee.
Folasade May: Exact Sciences – Consultant, Grant/Research Support. Freenome – Consultant. Geneoscopy – Consultant.
Julia J.. Liu, MD1, Samir Gupta, MD, MSCS2, Aasma Shaukat, MD3, Nik Sheng Ding, MBBS, PhD4, Heiko Pohl, MD5, Khushbu Desai, MS6, Dharini Chandrasekar, DO, MPH6, Christina Guerrier, MBA6, Prasanna Ramachandran, PhD6, Karina Islas-Rios, BSc, PhD7, Richard D. Shipman, BSc8, Tomislav Čaval, BSc, MSc, PhD6, Matthew P. Campbell, PhD9, Chirag Dhar, MD, MAS10, Gege Xu, PhD6, Flavio Schwarz, PhD6, Daniel Serie, MS6, Evelien Dekker, MD, PhD11, Michael B. Wallace, MD, MPH12, Daniel Hommes, MD, PhD6, Folasade P.. May, MD, PhD, MPhil13. P0359 - Targeting the Sugar Code: Serum Glycoproteome Profiling for Early Detection of Advanced Adenoma and Colorectal Cancer, ACG 2023 Annual Scientific Meeting Abstracts. Vancouver, BC, Canada: American College of Gastroenterology.

