Tuesday Poster Session
Category: IBD
P3627 - Using Unsupervised Natural Language Processing to Automatically Identify Colonic Dysplasia in Pathology Reports From Patients With Chronic Colitis
Tuesday, October 24, 2023
10:30 AM - 4:00 PM PT
Location: Exhibit Hall
Has Audio

Nathaniel J. Spezia-Lindner, MD
Baylor College of Medicine
Houston, TX
Presenting Author(s)
Nathaniel J. Spezia-Lindner, MD1, Scott Berger, MD1, Arash Maghsoudi, PhD1, Javad Razjouyan, PhD1, Manreet Kaur, MD2
1Baylor College of Medicine, Houston, TX; 2Mayo Clinic Arizona, Scottsdale, AZ
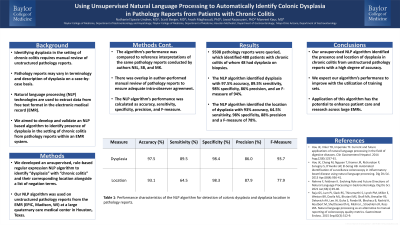
Introduction: Identifying dysplasia in the setting of chronic colitis requires manual review of unstructured pathology reports, which may vary in terminology or description of dysplasia. Natural Language Processing (NLP) technologies are used to extract data from free text format in the electronic medical record (EMR). We aimed to develop and validate an NLP based algorithm to identify presence of dysplasia in the setting of chronic colitis from pathology reports within an integrated EMR system.
Methods: We developed an unsupervised, rule-based regular expression NLP algorithm to identify “dysplasia” with “chronic colitis” and their corresponding location alongside a list of negation terms within pathology reports derived from the EMR (EPIC) at a large quaternary care medical center in Houston, Texas. The algorithm’s performance was evaluated in comparison to authors NSL, SB, and MK's interpretation of the contents of the same pathology reports. A portion of the pathology reports were reviewed by multiple authors to ensure adequate intra-observer agreement. The algorithm's performance was calculated as accuracy, sensitivity, precision and F- measure.
Results: We queried 9508 pathology reports and identified 480 patients with chronic colitis, of whom 48 had dysplasia on colonic biopsies. The NLP algorithm identified dysplasia with 97.5% accuracy, 89.5% sensitivity, 86% precision and an F-measure of 93.7% when compared with manual review. The NLP algorithm was able to identify the location of dysplasia with 93% accuracy, 87.9% precision and an F-measure of 78%.
Discussion: Unsupervised NLP approach identified the presence and location of dysplasia in the setting of chronic colitis with high degree of accuracy from pathology reports. We expect our algorithm’s performance to improve with the utilization of training sets. Application of this algorithm has the potential to improve patient identification to enhance research and clinical care across large EMRs.
Disclosures:
Nathaniel J. Spezia-Lindner, MD1, Scott Berger, MD1, Arash Maghsoudi, PhD1, Javad Razjouyan, PhD1, Manreet Kaur, MD2. P3627 - Using Unsupervised Natural Language Processing to Automatically Identify Colonic Dysplasia in Pathology Reports From Patients With Chronic Colitis, ACG 2023 Annual Scientific Meeting Abstracts. Vancouver, BC, Canada: American College of Gastroenterology.
1Baylor College of Medicine, Houston, TX; 2Mayo Clinic Arizona, Scottsdale, AZ
Introduction: Identifying dysplasia in the setting of chronic colitis requires manual review of unstructured pathology reports, which may vary in terminology or description of dysplasia. Natural Language Processing (NLP) technologies are used to extract data from free text format in the electronic medical record (EMR). We aimed to develop and validate an NLP based algorithm to identify presence of dysplasia in the setting of chronic colitis from pathology reports within an integrated EMR system.
Methods: We developed an unsupervised, rule-based regular expression NLP algorithm to identify “dysplasia” with “chronic colitis” and their corresponding location alongside a list of negation terms within pathology reports derived from the EMR (EPIC) at a large quaternary care medical center in Houston, Texas. The algorithm’s performance was evaluated in comparison to authors NSL, SB, and MK's interpretation of the contents of the same pathology reports. A portion of the pathology reports were reviewed by multiple authors to ensure adequate intra-observer agreement. The algorithm's performance was calculated as accuracy, sensitivity, precision and F- measure.
Results: We queried 9508 pathology reports and identified 480 patients with chronic colitis, of whom 48 had dysplasia on colonic biopsies. The NLP algorithm identified dysplasia with 97.5% accuracy, 89.5% sensitivity, 86% precision and an F-measure of 93.7% when compared with manual review. The NLP algorithm was able to identify the location of dysplasia with 93% accuracy, 87.9% precision and an F-measure of 78%.
Discussion: Unsupervised NLP approach identified the presence and location of dysplasia in the setting of chronic colitis with high degree of accuracy from pathology reports. We expect our algorithm’s performance to improve with the utilization of training sets. Application of this algorithm has the potential to improve patient identification to enhance research and clinical care across large EMRs.
Disclosures:
Nathaniel Spezia-Lindner indicated no relevant financial relationships.
Scott Berger indicated no relevant financial relationships.
Arash Maghsoudi indicated no relevant financial relationships.
Javad Razjouyan indicated no relevant financial relationships.
Manreet Kaur indicated no relevant financial relationships.
Nathaniel J. Spezia-Lindner, MD1, Scott Berger, MD1, Arash Maghsoudi, PhD1, Javad Razjouyan, PhD1, Manreet Kaur, MD2. P3627 - Using Unsupervised Natural Language Processing to Automatically Identify Colonic Dysplasia in Pathology Reports From Patients With Chronic Colitis, ACG 2023 Annual Scientific Meeting Abstracts. Vancouver, BC, Canada: American College of Gastroenterology.
